Genomic reassortments: a major source of influenza pandemics
PDFInfluenza viruses belong to the family Orthomyxoviridae, characterized by a segmentation of their RNA genome. As in all viruses with an RNA genome, mutation is an important source of diversity in these viruses, due to errors in the replication of their enzyme (RNA polymerase). Viruses with segmented genomes, such as the influenza virus, have another mechanism for generating diversity: reassortment.
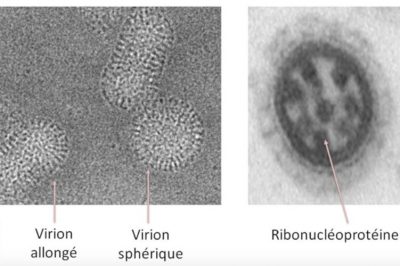
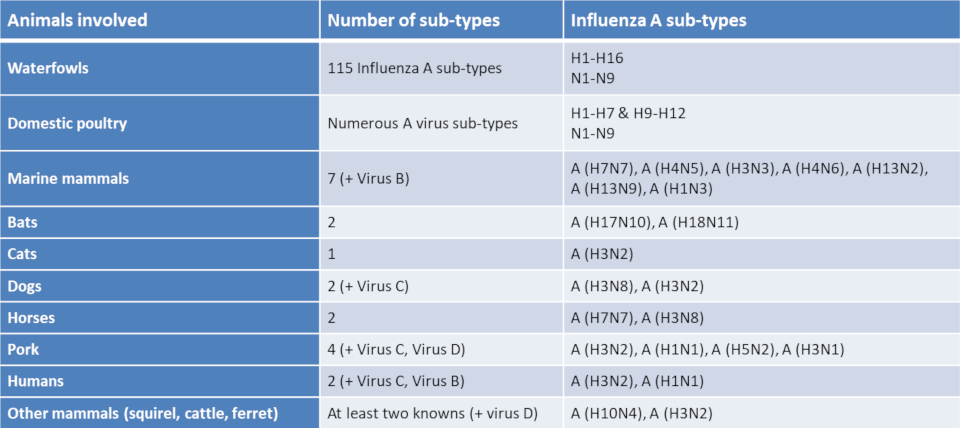
Figure 1 shows the structure of these viruses enveloped by two proteins: haemagglutinin H and neuraminidase N. Inside the viral particle appear the 8 RNA segments surrounded by a nucleoprotein, the whole being called “ribonucleoproteins”. There are four types of influenza viruses (also called Influenza Virus), A, B, C and D :
- Types A and B are the main types that spread regularly in humans and cause seasonal influenza epidemics.
- Influenza B almost exclusively infects humans and is less common than influenza A. Mutations are about two to three times slower than those of influenza A. Since humans are the natural host for influenza B, influenza B viruses do not create a pandemic.
- Type C viruses only cause mild respiratory infections.
The World Health Organization (WHO) has classified influenza A viruses into subtypes based on the surface proteins H and N, which respectively control the entry and exit of viruses into and out of attacked cells (see table).
Table. Classification of influenza A viruses into subtypes based on surface proteins H and N according to the World Health Organization (WHO).
When a strain of influenza A virus infects a cell, each RNA segment migrates into the nucleus. There, they are copied several times to form RNA genomes for new infectious virions. The new RNA segments are exported to the cytoplasm and then incorporated into new viral particles that bud from the cell.
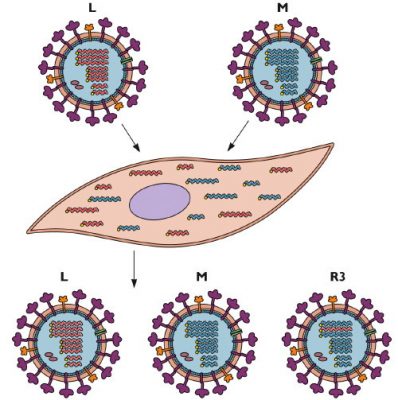
If a cell is infected with two different virus strains, the RNAs of both viruses are copied into the nucleus. When new virus particles are assembled at the plasma membrane, each of the 8 RNA segments can come from either of the infecting viruses. The offspring that inherit RNA from both parents are called “reassortments”. This is the situation that can be seen in the lungs of pigs, whose cells have receptors for both avian and mammalian strains. This process is illustrated in the diagram below (Figure 2), which shows a cell that is co-infected with two influenza L and M viruses. The infected cell produces both parental viruses as well as a reassortant R3 that inherits a segment of RNA from the L strain and the rest of the M strain.
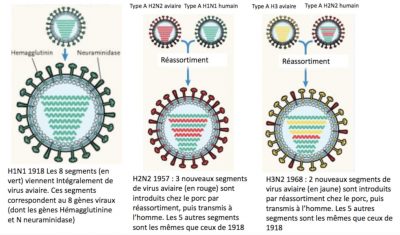
An example of the evolutionary importance of reassortment is the exchange of RNA segments between “mammalian” and “avian” influenza viruses that give rise to pandemic influenza. In 1957, 1968 and 2009, reassortment events in the intermediate host (pigs) led to new viruses, resulting in the Asian, Hong Kong and Mexican influenza pandemics. Figure 3 shows the reassortment sequences produced in pigs. The 1957 influenza virus (type A H2N2) acquired three genetic segments from an avian species (one coding for haemagglutinin, one coding for neuraminidase and one polymerase gene, PB1), and the 1968 influenza virus (type A H3N2) acquired two genetic segments from an avian species (haemagglutinin and PB1).
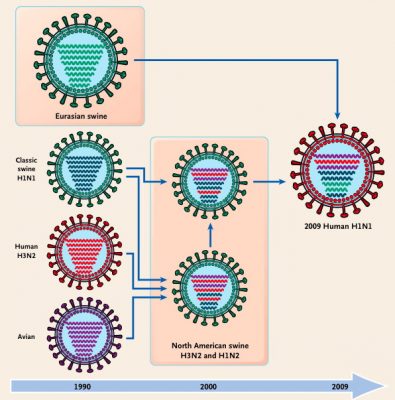
The pandemic H1N1 2009 strain is a reassortment of avian, human and swine influenza viruses, as shown in Figure 4. Reassortment can only occur between influenza viruses of the same type. It is not understood why influenza A viruses never exchange RNA segments with influenza B or C viruses. However, the reason is probably related to the viral packaging mechanism that ensures that each influenza virion contains at least one copy of each RNA segment.
In conclusion
These reassortment mechanisms, described as antigenic jumps, result in the emergence of radically new strains (at the antigenic level), making the immunity established in previous epidemics obsolete. Finally, in order for a pandemic influenza A virus to develop, it is necessary for such a reassortant virus to acquire, at a minimum, through mutations, the ability to recognize the human receptor to spread in the human population.
To find out more:
- Flu pandemics. Season I: What’s a pandemic?
- Influenza pandemics. Season II: How can an influenza pandemic occur?
- Trifonov, V., Khiabanian, H., & Rabadan, R. (2009). Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus New England Journal of Medicine DOI: 1056/NEJMp0904572