Ocean viruses
PDFLucie Bittner, Lecturer in bioinformatics and evolutionary genomics at Sorbonne Université, Paris
Chris Bowler, CNRS Research Director, Institut de Biologie de l’École Normale Supérieure, École Normale Supérieure, Paris
The Tara Oceans expedition has enabled the first large-scale study of viruses in the marine environment. Sequencing of 43 samples corresponding to 26 stations allowed to study the diversity and structure of viral communities between 6 oceanic provinces (i.e. Mediterranean Sea, Red Sea, western Indian Ocean, southern Atlantic Ocean, Southern Ocean, southern Pacific Ocean) [3].
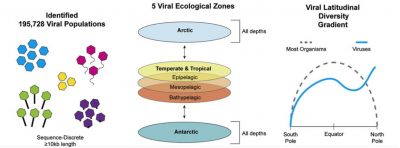
Metagenomic assembly of 145 marine viromes revealed 195,728 viral populations (Figure 1) [4]. Since viruses do not possess ribosomal genes that allow them to be classified taxonomically, it is the study of viral proteins that has made it possible to detect and define:
- A set of groups common to all samples or core group;
- The union of all detected clusters, corresponding to 1.3 million clusters (called pan-virome).
Metagenomic and morphological data reveal biogeographic patterns in which viral communities are passively transported by ocean currents, and locally structured by environmental conditions (Figure 1):
- The viral communities of the world ocean can be separated into five distinct ecological zones, including two distinct Arctic regions;
- Viral macrodiversity (inter-population diversity) and microdiversity (intra-population diversity) do not follow the latitudinal variation in diversity.
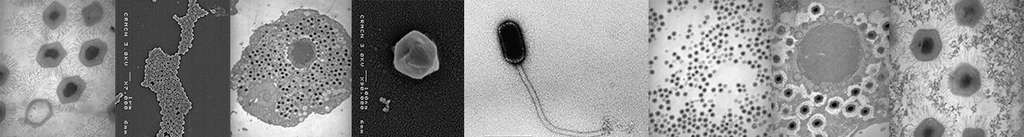
Quantitative morphological data obtained by transmission electron microscopy complement these observations (Figure 2).
- mortality;
- horizontal gene transfer [5];
- modulation of the metabolism of microbial communities.
Viruses target a variety of hosts both prokaryotic, such as the cyanobacterium Synechococcus, and eukaryotic such as the protist Emiliania huxleyi. They play an important role in the evolution of their hosts [6].
This new understanding of oceanic viruses is essential for their wider integration into ecosystem models. Thus, viruses would play an important role as “regulators” of planktonic populations but also of matter flows. For example, by lysing phytoplanktonic organisms that fix ambient carbon via photosynthesis, viruses contribute to the flow of carbon from the upper layers of the ocean to the depths, and even into the sediments (See: Focus: The ocean’s biological carbon pump).
Notes and references
[1] Giant viruses were discovered at the turn of the century in a variety of environments. They were so called because they are larger than other viruses both in particle size and genome length. These giant viruses, such as mimiviruses, pandoraviruses and jellyfish viruses isolated from cooling towers, ponds, hot springs and the ocean, add new mysteries to the origin and evolution of life.
[2] Breitbart M., 2012, Marine viruses: truth or dare. Annu Rev Mar Sci. 4:425-448; Fischer M.G. et al., 2010, Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. USA, 107:19508-19513.
[3] Gregory A.C. et al. (2019). Marine DNA viral macro- and microdiversity from Pole to Pole. Cell. 177(5):1109-1123.e14. doi: 10.1016/j.cell.2019.03.040.
[4] Fuhrman J. A., 1999, Marine viruses and their biogeochemical and ecological effects. Nature, 399:541-48; Weitz J.S. & Wilhelm S.W., 2012, Ocean viruses and their effects on microbial communities and biogeochemical cycles,” F1000 Biology Reports, 4(17).
[5] Ignacio-Espinoza J.C. & Sullivan M.B., 2012, Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes.” Environ Microbiol14(8): 2113-2126.
[6] Koskella B. & Brockhurst M.A., 2014, Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities.” Fems Microbiology Reviews, 38(5):916-931; Frada M., et al., 2008, The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc. Natl. Acad. Sci. USA, 105(41):15944-15949.




