Antibiotics, antibiotic resistance and environment
PDF
Microorganisms naturally produce antibiotics that inhibit the growth of other competing microorganisms. To survive, they develop genetic and biochemical mechanisms of antibiotic resistance, which have spread widely between species due to their frequent genetic exchanges. However, since the 1940s, humans have developed new antibiotics, and their use has become massive: in 15 years, global antibiotic consumption has increased by 65%, mainly in middle- and low-income countries. This global phenomenon is now a major public health concern. Some bacteria that are pathogenic to humans and animals have become resistant to most of the antibiotic molecules developed by the pharmaceutical industry. In addition, antibiotics administered to humans and animals, and resistant bacteria selected from these hosts, have been widely disseminated in the environment. Bacteria have also evolved towards an increase in their antibiotic resistance capacities. There are permanent exchanges between bacterial flora in environmental, animal or human reservoirs. The fight against antibiotic resistance must therefore be considered comprehensively. Shouldn’t it take into account the effects of antibiotics on these different reservoirs?
- 1. Antimicrobial resistance in bacteria: A global health threat
- 2. The environment: an old antibiotic resistance reservoir
- 3. Release of human-made antibiotics into the environment
- 4. Release of novel antibiotic resistance genes of human and animal origin into the environment
- 5. Emergence of new antibiotic-resistant products in the environment
- 6. The environment as a source of new antibiotic resistance in humans and animals
- 7. Synthesis and perspectives: A better understanding of the risks associated with the environment and intensive livestock farming
- 8. Messages to remember
1. Antimicrobial resistance in bacteria: A global health threat
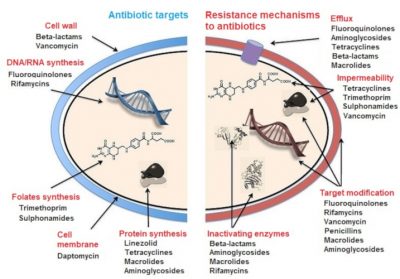
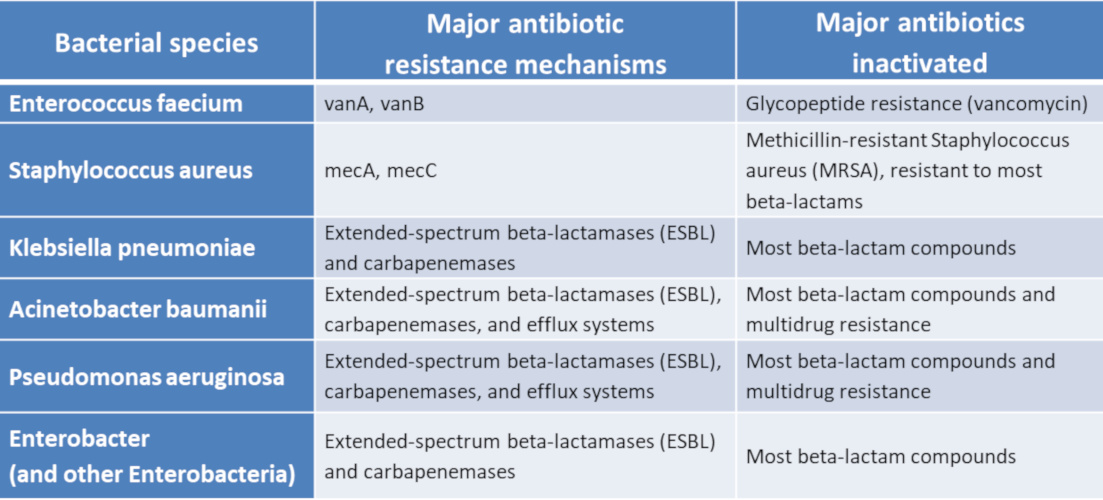
- the impermeability of the bacterial membrane to this antibiotic;
- the efflux of the antibiotic out of the bacterium after penetration;
- the reduction of the antibiotic’s affinity for its bacterial target(s), by quantitative or qualitative modification of the latter;
- and the inactivation of the antibiotic by bacterial enzymes.
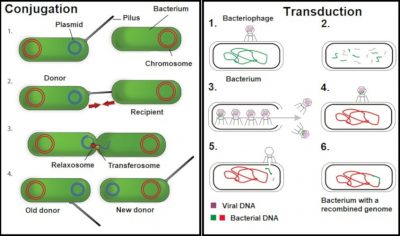
In some bacterial species, these antimicrobial resistance mechanisms are stable and affect most strains: we speak of natural antimicrobial resistance. On the contrary, acquired antibiotic resistance occurs when certain strains of a bacterial species usually susceptible to an antibiotic acquire a genetic mechanism of resistance to it. The acquisition of new antibiotic resistance may be linked to mutations (see Genetic Polymorphism and Variation) affecting the properties or level of expression of resistance genes. It may also be the consequence of the acquisition of new resistance genes. Indeed, mobile genetic elements [1] (plasmids, transposons, integrons, etc.) can be exchanged between bacteria of the same species or of different species. These exchanges take place through horizontal gene transfer mechanisms [2] (conjugation, transformation, transduction, transposition) (Figures 2 and 3). All antibiotic resistance genes present in pathogenic or non-pathogenic microorganisms constitute the resistome.
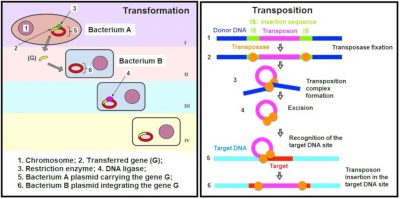
Antimicrobial resistance is currently a major global public health problem that has required the development of international (including WHO) and national control plans. While the human and animal reservoirs of these antibiotic resistances are well characterized, the role of the environment in their emergence and spread has only recently been recognized.
2. The environment: an old antibiotic resistance reservoir
Many environmental microorganisms (mainly bacteria and fungi) naturally produce antibiotics, which give them a selective growth advantage over other environmental species. These microorganisms possess genes encoding antibiotic resistance mechanisms to the antibiotics they produce. They can transmit these resistance genes to their offspring (vertical transmission) [4]. Microorganisms that do not produce antibiotics may also naturally harbour antibiotic resistance genes or acquire them de novo. In bacteria, there are many genetic exchanges between individuals of the same or different species (horizontal transmission). The spread of antimicrobial resistance between bacteria living in the environment is probably very old (several billion years) and continues today [5]. These resistances can be transmitted from environmental bacteria to those that colonize humans and animals. Examples include the recent emergence of extended-spectrum beta-lactamases (ESBLs) of the CTX-M type or a new mechanism for resistance to fluoroquinolones (qnr genes) [6].
3. Release of human-made antibiotics into the environment
The beginning of the 20th century was marked by the discovery and especially the mass production of natural antibiotics (extracts of microorganisms that produce these molecules) or obtained by chemical synthesis. Natural antibiotics include beta-lactam antibiotics (including penicillin), aminoglycosides, tetracyclines, and macrolides. Most of these natural molecules have been chemically modified to become semi-synthetic and more recently synthetic antibiotics. Sulphonamides and quinolones are molecules that have been immediately synthesized by humans. From the 1940s to nowadays, antibiotic use in humans and animals has increased exponentially, corresponding to the release of millions of tons of antibiotics into the environment. There are three main sources of environmental pollution by antibiotics:
- industrial antibiotic production plants that disperse part of this production in their aquatic environment;
- the breeding of domestic animals treated with antibiotics;
- humans who receive antibiotics.
3.1. Antibiotic production plants
Antibiotic production plants release large quantities of antibiotics in their effluents. Current regulatory constraints in terms of environmental pollution are insufficient in countries producing large quantities of antibiotics, particularly in Europe, the United States, China, and India. Antibiotic concentrations that may exceed 1 mg/L have been detected in effluents from some production plants [7]. These concentrations have a significant impact on the environmental microbial flora. Also, these antibiotics spread widely in the environment and pollute groundwater.
3.2. Veterinary use of antibiotics
Antibiotic use in domestic animals (wild animals are rarely involved) accounts for about 60% of total antibiotic production and continues to increase [8]. More than 100,000 tons of antibiotics are administered annually worldwide to livestock, lowland animals and aquaculture [9]. Tetracyclines in particular, followed by penicillins and sulfonamides, are the most commonly used antibiotics. However, beta-lactam antibiotics, aminoglycosides, phenicols, macrolides and glycopeptides are also concerned. Antibiotics used in humans and animals belong to the same families and are therefore inactivated by the same antimicrobial resistance mechanisms. Antibiotics are administered to domestic animals to treat infectious diseases, to prevent them (antibiotic prophylaxis), or as food supplements. For the treatment and prophylaxis of bacterial infections, antibiotics are often dispensed in drinking water or feed to the entire farm, even if only a few animals are sick. The systematic use of antibiotics as feed supplements (especially livestock, poultry, fish, etc.) is intended to improve livestock productivity (in terms of the amount of meat produced, for example).
This use of antibiotics for profitability purposes has been the overwhelming majority for several decades. It is currently banned in Europe, but its use persists in many countries. It is recognized as a major cause of antibiotic resistance in bacteria (e. g. Escherichia coli) colonizing the digestive tract of these animals. Most of the resistant bacteria selected in these animals can then be transmitted to humans through contact with them or through contaminated food products. Besides, antibiotics administered to animals are present in an active form in their feces. Wastewater from these farms is often directly released into the aquatic environment or used to irrigate crops without prior treatment.
3.3. Antibiotic use in humans
Antibiotics are used in humans, either to treat infectious diseases or to prevent them (antibiotic prophylaxis). Global human consumption of antibiotics corresponds to several tens of billions of unit doses each year. Beta-lactam antibiotics (penicillins, cephalosporins, carbapenems) are currently the most widely used antibiotics (about 60% of human consumption). The use of tetracyclines, macrolides, and fluoroquinolones is also high. About 80% of antibiotics are used in the community and 20% in hospitals. However, the role of health care institutions in the spread of bacterial resistance is major due to the broad spectrum of antibiotics used and the high risk of human-to-human transmission of selected resistant bacteria [10]. Some of the antibiotics prescribed to infected people are not used and are therefore released with our daily waste into the environment.
More significantly, most antibiotics absorbed by humans are eliminated in active form in urine and faeces. In developing countries, these antibiotics are directly dispersed in the hydro-telluric environment. In countries with wastewater treatment infrastructures, a large proportion of these antibiotics are found in wastewater treatment plants [11]. Concentrations in the order of µg/L of antibiotics (beta-lactam antibiotics, macrolides, tetracyclines, fluoroquinolones, etc.) have been detected in wastewater from some treatment plants. These antibiotics are biodegraded, absorbed in sewage sludge, or eliminated unchanged in the plant effluent. Sewage sludge can be used as fertilizer in crop fields, while effluent is discharged into the aquatic environment (rivers). In both cases, the active antibiotics are then released into the hydro-telluric environment.
- Residual concentrations of antibiotics in the environment
Antibiotics are detected in some aquatic and terrestrial environments at concentrations ranging from ng to µg per litre of water or per gram of soil. Similar concentrations have also been measured in some groundwater. These antibiotics persist in the environment after contamination for periods that can range from a few days (e.g., beta-lactam antibiotics) to several months (e.g., fluoroquinolones and tetracyclines). Therefore, for the latter molecules, there is an accumulation phenomenon if environmental contamination is permanent or frequent. Antibiotic residues can also be detected in the marine environment. Finally, residual concentrations of antibiotics in the order of ng/L have sometimes been detected in drinking water.
4. Release of novel antibiotic resistance genes of human and animal origin into the environment
Antibiotics administered to humans and animals select from their commensal flora (their microbiota) the bacterial species and strains most resistant to these molecules (see Human microbiots: Allies for our health).
Antibiotics also promote gene mutations and horizontal gene transfers encoding antibiotic resistance by induction in bacterial SOS systems. This phenomenon is particularly important in the intestinal microbiota, which is rich and contains several thousand bacterial species (Figure 4).
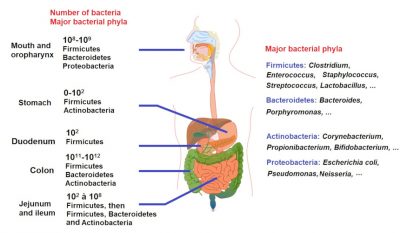
5. Emergence of new antibiotic-resistant products in the environment
The hydro-telluric environment is a medium rich in microorganisms whose diversity is only partially characterized. Concerning bacteria, recent metagenomic studies have led to the discovery of many new species. They also showed that more than 99% of environmental bacterial species are not cultivable with current methods [12].
This environment is increasingly polluted by bacteria resistant to antibiotics of human or animal origin, by genes encoding these resistances, and by human-made antibiotics. Plants absorb some of these antibiotics, particularly in crops irrigated with water or enriched with sludge from wastewater treatment plants contaminated by these molecules. While residual antibiotic concentrations in the environment are usually low, they nevertheless exert a progressively increasing selection pressure on the natural microbial flora. This selection pressure promotes horizontal exchanges of antibiotic resistance genes between natural and polluting species, particularly in the rich and varied bacterial populations located in biofilms. The natural predators of these microorganisms, the protozoa that are free-living organisms (amoebas in particular), also promote these genetic exchanges.
All in all, all the conditions are in place for new antibiotic resistance to emerge in the environment, both in the natural environmental bacterial flora, but also in the human and animal flora capable of surviving in this environment.
6. The environment as a source of new antibiotic resistance in humans and animals
Microorganisms from their hydro-telluric environment frequently colonize humans and animals. This colonization can be direct through contact with this environment during daily activities or indirect through ingestion of contaminated drinking water or food, or exposure to soiled objects etc.) [13]. The commensal microbial flora of humans and animals acts as a microbial barrier that protects against infection by less well adapted exogenous species. Nevertheless, many human and animal bacterial species survive in the environment and can, therefore, re-contaminate these hosts. On the other hand, environmental reservoir microorganisms can colonize the skin and mucous membranes of humans and animals. This colonization can be transient and of short duration (a few hours or days) or prolonged (a few weeks to several months or even years). During this period of colonization, genetic exchanges between this exogenous flora and the endogenous commensal flora can occur. These exchanges are particularly important at the level of the digestive microbiota, due to the richness and diversity of this microbiota, but also the prolonged duration of intestinal carriage of many environmental microorganisms after their ingestion.
It should be noted that antibiotic-resistant bacteria are also often resistant to antiseptics. These include heavy metals (silver, copper, mercury, etc.) and biocides (ethanol, formaldehyde, chlorhexidine, triclosan, quaternary ammonium, etc.). Environmental pollution by antiseptics can contribute to the selection of microorganisms that carry antibiotic resistance. Two phenomena make it possible to understand that an antiseptic selects strains carrying antibiotic resistance (or that an antibiotic selects for resistance to an antiseptic):
- the genetic mechanisms of resistance to an antibiotic and an antiseptic are carried by the same genetic carriers (plasmids in particular); exposure of a bacterium to the antibiotic or antiseptic concerned selects the strains carrying resistance to both types of compounds: it is co-resistance ;
- the same genetic mechanism (encoding an efflux pump, for example) leads to resistance to both an antibiotic and an antiseptic; it is the phenomenon of cross-resistance that leads to the selection of both types of resistance in the presence of one or the other of these molecules.
7. Synthesis and perspectives: A better understanding of the risks associated with the environment and intensive livestock farming
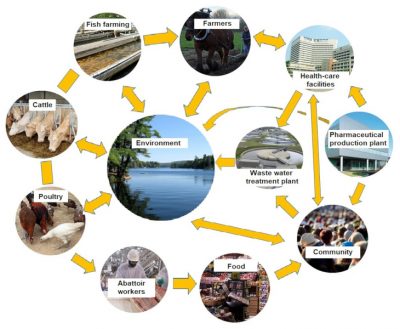
Antimicrobial resistance is a complex and long-standing phenomenon, which has become a major problem for human and animal health due to the combined effects of three actors: 1/ overconsumption of antibiotics for several decades; 2/ high genomic plasticity and therefore adaptability of bacteria to the selection pressure exerted by these molecules; and 3/ permanent exchanges between the three major reservoirs of resistance genes: the environment, animals and humans (Figure 5).
The numerous and rapid international trade (human, animal, food) has also aggravated this situation. The environment is polluted by both resistant microorganisms of human and animal origin, and by residual concentrations of antibiotics. These two types of pollution promote the emergence of new antibiotic resistance and their transfer between bacterial species.
Due to the very strong interactions between human, animal and environmental reservoirs, the spread of genetic mechanisms of antimicrobial resistance is a rapid and major phenomenon. Compared to other environmental pollutants (other drugs, chemicals, heavy metals, etc.), this pollution is characterized by its possible spread among the human and animal population. Indeed, the inter-human, inter-animal, and human-animal transmissions of antibiotic-resistant bacteria explain the individual but also the collective effects of this pollution. Among the means of combating the emergence and spread of antimicrobial resistance, it is essential in particular:
- to reduce and optimize overall antibiotic consumption;
- to avoid or at least limit the spread of antibiotics and bacteria carrying resistance genes in the environment, in particular from significant sources (hospitals, antibiotic manufacturing plants, intensive livestock farming, water treatment plants, etc.);
- to improve the level of hygiene and develop water purification systems in developing countries;
- to better monitor environmental pollution by antibiotics and antibiotic resistance genes [14], [15].
8. Messages to remember
- Global consumption of antibiotics is in the billions of daily doses and is increasing.
- Antibiotics consumed by humans and animals profoundly modify their commensal microbial flora and select bacterial strains that are increasingly resistant to these molecules.
- The adaptive capacities of human and animal pathogenic bacteria linked to their genomic plasticity allow them to develop and exchange new antimicrobial resistance mechanisms continually.
- Antibiotics consumed by humans and animals, as well as bacteria carrying antimicrobial resistance mechanisms, are dispersed in the environment in active form.
- The environment is a natural reservoir of many bacterial species and antibiotic resistance genes, enriched continuously by pollution from human and animal sources.
- Permanent exchanges between human, animal and environmental reservoirs perpetuate the emergence and spread of antibiotic resistance
- It is necessary to develop new antibiotics and new therapeutic strategies to combat antibiotic resistance, but also to reduce and optimize the consumption of current antibiotics in humans and animals.
References and notes
Cover image. Isolation of a bacterial colony on a selective medium, before testing for antibiotic sensitivity. [Source: Getty Images, royalty-free.]
[1] A mobile genetic element is a segment of the genome whose position is not fixed. These elements were discovered by the American cytogeneticist Barbara McClintock, who won the Nobel Prize in Physiology or Medicine in 1983. There is a wide variety of them, including plasmids, transposons, integrons.
[2] A process in which an organism integrates genetic material from another organism without being its descendant (also called lateral gene transfer). A large part of genetic engineering consists of the artificial horizontal transfer of genes.
[3] Pendleton JN, Gorman SP, Gilmore BF. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 11(3):297‑308.
[4] Finley RL, Collignon P, Larsson DGJ, McEwen SA, Li X-Z, Gaze WH, et al (2013). The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis, 57(5):704-10.
[5] D’Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, et al (2011). Antibiotic resistance is ancient. Nature. 477(7365):457‑61.
[6] Cantón R. (2009). Antibiotic resistance genes from the environment: a perspective through newly identified antibiotic resistance mechanisms in the clinical setting. Clin Microbiol Infect. 15 Suppl 1:20-5.
[7] Larsson DGJ. (2014). Antibiotics in the environment. Ups J Med Sci. 119(2):108-12
[8] Singer AC, Shaw H, Rhodes V, Hart A. (2016). Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Microbiol Front. 7:1728.
[9] Lekshmi M, Ammini P, Kumar S, Varela MF. (2017). The Food Production Environment and the Development of Antimicrobial Resistance in Human Pathogens of Animal Origin. Microorganisms. 5(1).
[10] Hosein IK, Hill DW, Jenkins LE, Magee JT. (2002). Clinical significance of the emergence of bacterial resistance in the hospital environment. J Microbiol application, 92 Suppl:90S-7S.
[11] Le-Minh N, Khan SJ, Drewes JE, Stuetz RM. (2010). Fate of antibiotics during municipal water recycling treatment processes. Water Res. 44(15):4295-323.
[12] Nesme J, Cécillon S, Delmont TO, Monier J-M, Vogel TM, Simonet P. (2014). Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol, 24(10):1096-100.
[13] Zurek L, Ghosh A. (2014). Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl About Microbiol. 80(12):3562-7.
[14] Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al (2013). Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. About Health Perspect. 121(8):878‑85.
[15] Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, et al (2013). Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 13(12):1057-98.
The Encyclopedia of the Environment by the Association des Encyclopédies de l'Environnement et de l'Énergie (www.a3e.fr), contractually linked to the University of Grenoble Alpes and Grenoble INP, and sponsored by the French Academy of Sciences.
To cite this article: MAURIN Max (May 7, 2019), Antibiotics, antibiotic resistance and environment, Encyclopedia of the Environment, Accessed April 20, 2024 [online ISSN 2555-0950] url : https://www.encyclopedie-environnement.org/en/health/antibiotics-antibiotic-resistance-and-environment/.
The articles in the Encyclopedia of the Environment are made available under the terms of the Creative Commons BY-NC-SA license, which authorizes reproduction subject to: citing the source, not making commercial use of them, sharing identical initial conditions, reproducing at each reuse or distribution the mention of this Creative Commons BY-NC-SA license.