RubisCO
PDF1. From “Fraction 1 Protein” to RubisCO
Sam Wildman [1] was the first to characterize this protein around the 1950’s. [2] The abundance of RubisCO in leaf chloroplasts of C3 type plants (30-50% of proteins), and its high molecular weight (550 kDa) explain why the fractionation of leaf extracts using ammonium sulphate led to the preparation of a homogeneous protein fraction called “Fraction 1” representing a significant part of leaf proteins. Known as “Fraction 1 protein” for convenience, the content of this fraction has been extensively characterized by electrophoresis, analytical centrifugation, crystallization, etc., and has been used in a wide range of applications.
But its enzymatic function remained a mystery. It was discovered by another route. After deciphering the Benson-Bassham-Calvin Cycle, researchers set out to find the enzyme that catalyzes the binding of CO2 to Ribulose 1,5-diphosphate followed by the dismutation of 3-PGA into two molecules (see Focus Deciphering the Benson-Bassham-Calvin Cycle). Discovered in 1956 [3] this enzyme was called carboxydismutase. It then took only a short time to demonstrate that the enzyme thus identified and the “Fraction 1 protein” were one and same protein: Ribulose bisphosphate (or RuBP) Carboxylase.
Then, in the 1970s, the ability of ribulose biphosphate carboxylase to also bind oxygen was demonstrated. [4] This enzyme is therefore bifunctional and exerts in addition to its carboxylase activity a second activity called oxygenase, hence the name RubisCO (Ribulose biphosphate Carboxylase Oxygenase).
RubisCO, an enzyme specific to photosynthesis, has been the gateway for carbon to enter most of the planet’s organic molecules for more than three billion years. It is also at the origin of photorespiration, a metabolic pathway that appeared much later, with the increase in the oxygen content of the atmosphere. It is the most quantitatively important enzyme in the biosphere. In addition, it is the main reserve of organic nitrogen in the leaves.
2. Structure of the RubisCO
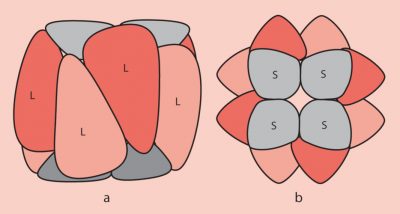
- The large subunits are encoded in the chloroplast stroma by the chloroplast genome ;
- The small subunits are encoded by the nuclear genome of the photosynthetic cells and are then imported into the chloroplast stroma through the outer and inner membranes of the chloroplast organelle;
- The folding of the polypeptides corresponding to the subunits and their assembly to form the functional RubisCO involves chaperone proteins ;
- At the functional level, the large sub-units carry the catalytic sites. The small sub-units, necessary for its operation, have a regulatory role.
3. Activity of the RubisCO
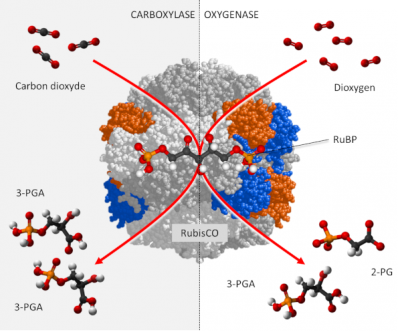
RubisCO is a carboxylase [6], which ensures – within the chloroplast stroma – the fixation of the carbon of CO2 on a phosphorylated sugar with 5 carbon atoms, RuBP or Ribulose 1,5-bisphosphate (see Focus Deciphering the Benson-Bassham-Calvin Cycle), to form an unstable molecule with 6 carbon atoms which will quickly give two molecules of 3-phosphoglyceric acid (3-PGA) with 3 carbon atoms according to the reaction:
RuBP + CO2 → 2 3-PGA (carboxylation reaction)
In addition, carbon dioxide and oxygen are competing at the RubisCO catalyst sites:
- In the presence of a high concentration of CO2, RubisCO functions only as a carboxylase leading to the synthesis of PGA molecules reduced to phosphate trioses, the origin of the phosphorylated sugars formed by the Benson-Bassham-Calvin cycle.
- On the other hand, in the presence of a high concentration of O2 and a low concentration of CO2, RubisCO gives rise to a PGA molecule and a two-carbon molecule, phosphoglycolate (P-glycolate), which is rapidly dephosphorylated to glycolate.
RuBP + O2 → PGA + P-glycolate (oxygenation reaction)
In fact, CO2 and O2 are involved in two antagonistic activities (see The path of carbon in photosynthesis). (Figure 2) :
- carbon dioxide promoting the carboxylase function of the RubisCO ;
- oxygen promoting the oxygenase function manifested by photorespiration.
Prior to these reactions, the RubisCO must be modified in its conformation by a RubisCO activase in the presence of ATP, and activated by CO2 in the presence of magnesium.
RubisCO is not a very active enzyme: the value of its average turnover (or catalytic constant) is between 1 and 10 s-1. This means that each molecule of RubisCO can, depending on the conditions, produce between one and 10 molecules of G3P per second, which is low. [7] This is the limiting stage of the Benson-Bassham-Calvin Cycle. It can therefore become a limiting factor in photosynthesis, even under favourable conditions.
The low activity of the RubisCO has mechanical and evolutionary reasons. RubisCO evolved prior to the first major oxygenation event in an oxygen-free atmosphere so that its functional mechanism was not constrained by the presence of this gas. When the oxygen content of the atmosphere increased, the RubisCO had to distinguish between carbon dioxide and oxygen. However, the more specific an enzyme is, the slower its catalytic function. To better discriminate between oxygen and carbon dioxide, the RubisCO has therefore reduced its reaction speed. [8]
4. Origin of the RubisCO
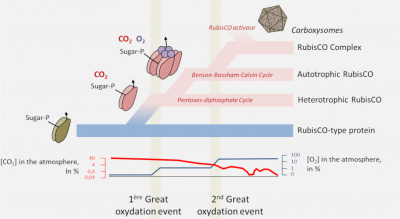
- The proteins of the RPL/RubisCO superfamily probably originate from a non-CO2-binding enolase-type enzyme. [9] It is within this family that a true carboxylase would have appeared;
- RubisCO would have appeared in archaea in a non-autotrophic context from nucleotide metabolism before RubisCO-dependent autotrophy appeared in bacteria with the Benson-Bassham-Calvin cycle;
- In order to optimize the carbon flow RubisCO would then be transformed from an autonomous enzyme to an enzyme complex, the ultimate evolution being found in cyanobacteria with the carboxysomes or pyrenoids of algae.
Notes and References
Cover image. Structure of RubisCO (Public domain)
[1] Samuel Goodnow Wildman (1912-2004), American biologist.
[2] Wildman S.G. & Bonner J. (1947) The proteins of green leaves. I. Isolation, enzymatic properties and auxin content of spinach cytoplasmic proteins. Arch. Biochem. 14:381-413; Singer S.J., Eggman L., Campbell J. & Wildman S.G. (1952) The proteins of green leaves. IV. High molecular weight protein comprising a large part of the cytoplasmic proteins. J. Biol. Chem. 197:233-239; Wildman S.G. (2002) Along the trail from Fraction I protein to Rubisco (ribulose bisphosphate carboxylase-oxygenase). Photosynth. Res. 73:243-250.
[3] Weissbach A., Horecker B.L. & Hurwitz J. (1956) The enzymatic formation of phosphoglyceric acid from ribulose diphosphate and carbon dioxide. J. Biol. Chem. 218: 795-810.
[4] Lorimer G.H. (1981). The carboxylation and oxygenation of ribulose 1,5-bisphosphate: The primary events in photosynthesis and photorespiration. Annu. Rev. Plant Physiol. 32: 349-383.
[5] Anderson I. & Backlund A. (2008) Structure and function of RubisCO. Plant Physiol Biochem. 46(3):275-91
[6] A carboxylase is an enzyme that binds mineral carbon (which is oxidized) to organic carbon (which is reduced) and thus ensures the incorporation of CO2 into molecules during carbohydrate synthesis. Carboxylation reactions are endergonic, i.e. they can only be carried out through the expenditure of energy (usually provided by the dephosphorylation of an adenosine triphosphate molecule)
[7] By way of comparison, one of the fastest enzymes – carbonic anhydrase – has a catalytic constant of up to 106 s-1, which means that each carbonic anhydrase molecule can produce up to one million bicarbonate ions per second.
[8] Erb T.J. & Zarzycki J. 2018, A short history of RubisCO: the rise and fall (?) of Nature’s predominant CO2 fixing enzyme. Cur. Op. Biotech. 49, 100-107 – https://doi.org/10.1016/j.copbio.2017.07.017
[9] Enolase catalyzes the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP)




