DNA barcodes to characterize biodiversity
PDF
New high-throughput molecular biology techniques make it possible to identify species in our environment. By characterizing short fragments of DNA that persist in the environment, it is possible to inventory biodiversity within ecosystems from e.g., water, soil or faeces samples. Such biodiversity surveys can be used to detect the presence of discrete species, to reconstitute paleo-environments or food-webs. They also offer prospects for DNA traceability and authenticity control of food and cosmetic products..
1. A universal diagnostic tool
The identification of species is not a new concern, as it has always concerned all human societies that take organisms from their environment for various purposes (e.g., food, medicine, culture). Now, characterizing biodiversity in terms of species becomes a major scientific and societal challenge. Indeed, identifying species is a necessary prerequisite for understanding their interactions that determine the functioning, dynamics and evolution of ecosystems. This step is also necessary to conserve or restore biodiversity in a context of global change with regards to e.g., climate, land use change and urbanization.
The species have been and are still mainly described on the basis of morphological criteria, which are increasingly substituted by molecular criteria based in particular on DNA characterisation (see What is Biodiversity?). The latter are particularly relevant for studying groups in which morphology is difficult to access, such as microorganisms, or not very variable, such as nematodes.
The Barcode of Life [1] project started in the 2000s to provide a universal diversity diagnostic tool that can be used in many fields such as ecology, agronomy, customs regulations, etc. This initiative has helped to accelerate the description of biodiversity. It is based on the concept of DNA-barcoding, which relies on the comparison of standardized genetic data obtained from an individual to that of specimens referenced in collections and identified by taxonomistsTaxonomists, whose purpose is to describe living organisms and group them into entities called taxons in order to (a) identify them, then (b) classify them and (c) recognize them using dichotomous determination keys., for a quick and reliable identification of the species. For this purpose, the DNA barcode used corresponds to a standard DNA fragment which sequence is species-specific. For example, researchers involved in the Barcode of Life initiative have defined a region of the mitochondrial gene encoding for the cytochrome oxidase 1Subunit 1 of the respiratory chain enzyme complex (abbreviated as COX1). This subunit is encoded by the mitochondrial genome, unlike the majority of genes encoding cytochrome oxidase subunits (encoded by the nuclear genome). The use of COX1 sequences makes it possible to discriminate between the various animal species, with the exception of Cnidarians. as the reference fragment for identifying animal species.
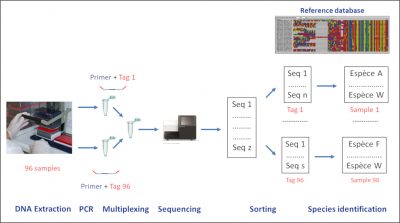
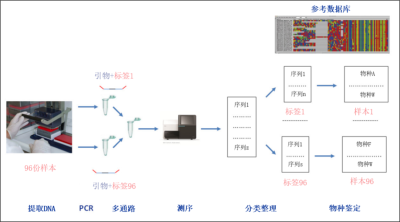
This concept of DNA-barcode is used by taxonomists, but also by ecologists who more broadly use any DNA fragment to identify specimens. This strategy makes it possible to identify the species present in an environment even though individuals are not easily accessible. This of course relates to micro-organisms, but also to animal and plant macro-organisms which presence can be detected thanks to the traces of DNA they leave through e.g., corpses, mucus or faeces. The principle is based on the extraction of the DNA from an environmental sample (water, soil, etc) followed by the PCRAbbreviation for “Polymerase Chain Reaction” or polymerase chain reaction. The English abbreviation has become common parlance. An in vitro targeted replication technique that produces, from a complex and sparse sample, large quantities of a specific, double-stranded DNA fragment of defined length. Each PCR cycle consists of three steps: DNA denaturation by heating to separate the two strands, hybridization of primers at the ends of the desired sequence, elongation by the action of a DNA polymerase, (Polymerase Chain Reaction) amplification of a target DNA region [2]. The PCR primersOligonucleotide sequences used in PCR reactions. They define, by limiting it, the sequence to be amplified. that can be species-specific for detecting a target species, for example an invasive species such as the bull frog, which has been detected even at low density from pond water samples. Conversely, primers can be defined in a way that is relevant to simultaneously study a wide range of species. This is called metabarcoding. In this case, the DNA fragments amplified during the PCR should be sequenced and compared to a reference base to link them to a given species (Figure 1).
2. A molecular biology approach for ecology
For a metabarcoding approach to effectively identify a species on the basis of its DNA present in an environmental sample, it is necessary to define convenient DNA barcodes. These barcodes must have a sequence variable between species but invariable within the same species, in order to have a high identification capacity (i.e., resolution). This sequence must be surrounded by two areas highly conserved from one species to another, to allow the simultaneous amplification of the target fragment in as many species as possible (i.e., large taxonomic coverage). In addition, the amplified fragment must be short to characterize degraded matrices. Indeed, the degradation of DNA in environmental matrices makes it difficult to amplify fragments longer than 150 nucleotides. In addition, since DNA is generally in a limiting quantity, the use of mitochondrial or chloroplastic DNA fragments is preferred as their copy number per cell is 100 to 1,000 times higher than that of nuclear DNA. It is also useful to define phylogenetically informative DNA barcodes (i.e., which divergence level reflects the divergence between species) in order to link unknown taxa to known related species. Data mining on large databases of DNA sequences with dedicated bioinformatics tools allow defining the most relevant barcodes for studying a group of organisms. These bioinformatics approaches also make it possible to evaluate the resolution and taxonomic coverage of these barcodes.

- The sampling must be done according to strict standards to avoid contaminations (Figure 2).
- The extraction of DNA from each sample is carried out according to a protocol adapted to the type of sample to be studied (water, soil, faeces…).
- The extracted DNA is then amplified by PCR with the primers targeting the barcode region. A short DNA label specific of each sample at one end of each primer is used as a tag to keep the information necessary to assign each sequence to its sample (Figure 1). At this stage, it is possible to block the amplification of a particular species by using an oligonucleotide binding specifically on the DNA of this species. For example, this is used during diet assessment from faeces, in order to block the amplification of the predator’s DNA, which is present in large quantities and could mask the detection of certain preys.
- After the PCR, each sample is represented by a mixture of the amplified DNA barcodes (amplicons) of the species it contains.
- The amplicons of the various samples are mixed during a multiplexing step (Figure 1).
- The High throughput sequencing of amplicons is performed. Next Generation Sequencing techniques have enabled the development of metabarcoding approaches that were previously unthinkable in practice. Current technologies allow several million DNA molecules to be sequenced in parallel [3].Starting from several hundred samples, a few thousand sequences per sample are sufficient to provide relevant information.
- After sequencing, the sequences are sorted by sample based on the tags and then assigned to species by comparison with reference sequences.
Throughout this process, bioinformatics tools are essential to sort the data, to build reference databases, to assign the sequences to the taxa via these databases, to define tag lists and manage sequencing errors.
Without species identification, the DNA sequences obtained can be used to define operational units (MOTUS for Molecular Operational Taxonomic Units), from which it is also possible to quantify the biodiversity of the samples. This is generally the case when working with microorganisms where most species are not described and not even cultivable.
3. Characterizing environmental samples: the metabarcoding approach
The metabarcoding approach offers alternatives to the more cumbersome methods that have been used to describe biodiversity up to now. It opens up new perspectives for studying the functioning and evolution of terrestrial and aquatic ecosystems, which require prior knowledge on the species communities interacting within them [4]. Describing biodiversity from soil samples using metabarcoding is useful, among other things, when individuals are difficult to find and identify morphologically. This is the case for many soil macro-fauna species which function within ecosystems is essential: earthworms, insects, springtails, etc. Metabarcoding can also replace traditional botanical surveys, particularly in environments where diversity is extremely high, such as tropical forests. The Amazonia contains 11,000 tree species, half of which are at risk of extinction. Traditional botanical methods would lead to ignoring up to 20% of the genera present; a much better resolution can be achieved by using DNA barcodes.
Finally, metabarcoding is especially effective in reconstructing paleo-ecosystems even though the species that made them up have disappeared. Conventional methods, such as the study of macro fossils and pollens, are cumbersome to implement and have a low taxonomic resolution. Early metabarcoding studies of permafrostGeological term that refers to a soil whose temperature remains below 0°C for more than two consecutive years. Represents more than 20% of the Earth’s surface. Permafrost is referred to in English and pergelisol in Russian. The permafrost is covered by a layer of soil, called an “active zone”, which thaws in summer and thus allows the development of vegetation. Its thawing under the effect of global warming has major consequences for the environment: methane release, release of pathogenic microorganisms, etc. (see The permafrost) samples dating back more than 20,000 years have shown a much better resolution than pollen analyses of samples. The study of different layers of Siberian pergelisolTerm used instead of permafrost for Siberia. (see Figure 2) showed that the steppes of the preglacial period were composed of forbs which were replaced by grasses during the postglacial period. In parallel, the analysis of fossilized stomach contents showed that mammoths, which disappeared after the glaciation, fed mainly on these forbs. The metabarcoding approach has thus made it possible to correlate the diet of an extinct animal with the drastic variation in the plant diversity of their ecosystem.
Thanks to the development of new sequencing techniques, metabarcoding has become an effective alternative to traditional methods for describing biodiversity from environmental samples [6]. In addition, it offers new perspectives for conducting integrated studies from the same sample, through the combined analysis of different barcodes. One can imagine the simultaneous analysis of the microbiotaAll microorganisms (bacteria, yeasts, fungi, viruses) living in a specific environment (called microbiome) in a host (animal or plant). An important example is the set of microorganisms living in the intestine or intestinal microbiota, formerly called “intestinal flora”. of the intestine or rumen, the procession of parasites and the diet of a species from its faeces. The scope of application is not limited to ecological studies, and extends to areas where the analysis of the composition of complex matrices via DNA characterization is an issue such as forensics, agri-food or customs controls.
References and notes
Cover image. [Source: © Jacques Joyard]
[1] http://www.barcodeoflife.org
[2] http://www.ens-lyon.fr/RELIE/PCR/principe/principe.htm
[3] https://www.ebi.ac.uk/training/online/course/ebi-next-generation-sequencing-practical-course/what-you-will-learn/what-next-generation-dna-
[4] Pompanon F, Coissac E, Taberlet P (2011) Metabarcoding, une nouvelle façon d’analyser la biodiversité. Biofutur, 319:30-32.
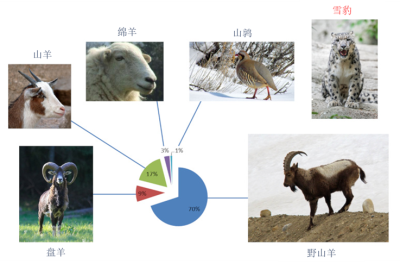
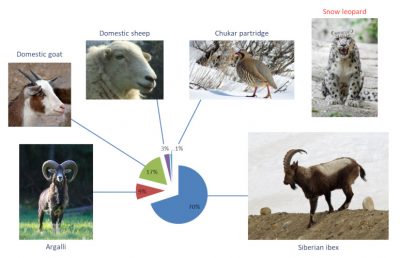
[5] Shehzad W et al (2012) Prey preference of snow leopard (Panthera uncia) in South Gobi, Mongolia. PLoS ONE 7(2): e32104. doi:10.1371/journal.pone.0032104
[6] Joly D, Faure D & Salamitou S (2015) Empreinte du vivant, l’ADN de l’environnement. Le Cherche Midi, 192 p.
The Encyclopedia of the Environment by the Association des Encyclopédies de l'Environnement et de l'Énergie (www.a3e.fr), contractually linked to the University of Grenoble Alpes and Grenoble INP, and sponsored by the French Academy of Sciences.
To cite this article: POMPANON François, SHEHZAD Wasim (April 26, 2019), DNA barcodes to characterize biodiversity, Encyclopedia of the Environment, Accessed July 27, 2024 [online ISSN 2555-0950] url : https://www.encyclopedie-environnement.org/en/life/dna-barcodes-to-characterize-biodiversity/.
The articles in the Encyclopedia of the Environment are made available under the terms of the Creative Commons BY-NC-SA license, which authorizes reproduction subject to: citing the source, not making commercial use of them, sharing identical initial conditions, reproducing at each reuse or distribution the mention of this Creative Commons BY-NC-SA license.